Farquhar et al. (1980) Photosynthesis
Table of contents
Code
Main program
sp_11_01.m | View on GitHub
% Supplemental program 11.1
% --------------------------------------------------------------------
% Calculate C3 photosynthesis in relation to PAR, CO2, and temperature
% --------------------------------------------------------------------
% --- Waveband indices for visible and near-infrared
params.vis = 1; params.nir = 2;
% --- Physical constants
physcon.tfrz = 273.15; % Freezing point of water (K)
physcon.rgas = 8.31446; % Universal gas constant (J/K/mol)
% --- Set leaf physiology variables
% Photosynthesis co-limitation: 0 = no. 1 = yes
leaf.colim = 1;
% Leaf physiological parameters
[leaf] = LeafPhysiologyParams (params, physcon, leaf);
% --- Plot type: 1 = light. 2 = CO2. 3 = temperature
plot_type = 1;
% --- Default conditions
co2conc = 380; % Atmospheric CO2 (umol/mol)
par_sat = 2000; % Incident PAR (umol photon/m2/s)
leaf_temp = physcon.tfrz + 25; % Leaf temperature (K)
% Atmospheric CO2 (umol/mol) and O2 (mmol/mol)
atmos.co2air = co2conc;
atmos.o2air = 0.209 * 1000;
% Leaf absorbed PAR (umol photon/m2 leaf/s)
atmos.par = par_sat;
flux.apar = atmos.par * (1 - leaf.rho(params.vis) - leaf.tau(params.vis));
% Leaf temperature (K)
flux.tleaf = leaf_temp;
% --- Light response at standard CO2 and leaf temperature
if (plot_type == 1)
p = 0;
for i = 0: 10: 2000
% Set value for PAR
atmos.par = i;
flux.apar = atmos.par * (1 - leaf.rho(params.vis) - leaf.tau(params.vis));
% Calculate photosynthesis
[flux] = LeafPhotosynthesis (physcon, atmos, leaf, flux);
% Save data for output
p = p + 1;
x1(p) = atmos.par;
x2(p) = flux.tleaf - physcon.tfrz;
x3(p) = atmos.co2air;
x4(p) = flux.ci / atmos.co2air;
x5(p) = flux.ac - flux.rd;
x6(p) = flux.aj - flux.rd;
x7(p) = flux.an;
end
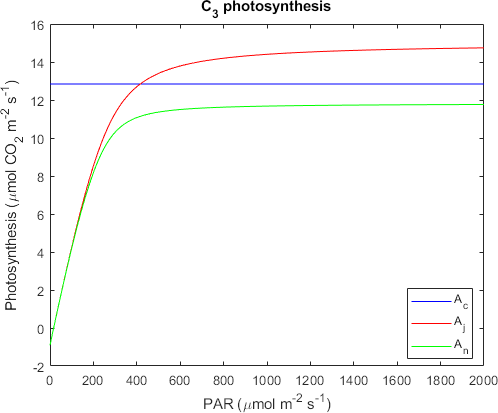
% Plot data
plot(x1,x5,'b',x1,x6,'r',x1,x7,'g')
title('C_3 photosynthesis')
xlabel('PAR (\mumol m^{-2} s^{-1})')
ylabel('Photosynthesis (\mumol CO_2 m^{-2} s^{-1})')
legend('A_c','A_j','A_n','Location','best')
% Output data and then clear variables from memory
A = [x1; x2; x3; x4; x5; x6; x7];
filename = 'light_response.txt';
fileID = fopen(filename,'w');
fprintf(fileID,'%9.4f %9.4f %9.4f %9.4f %9.4f %9.4f %9.4f\n', A);
fclose(fileID);
clear x1 x2 x3 x4 x5 x6 x7 A;
% Reset to default value
atmos.par = par_sat;
flux.apar = atmos.par * (1 - leaf.rho(params.vis) - leaf.tau(params.vis));
end
% --- CO2 response at saturated light and standard leaf temperature
if (plot_type == 2)
p = 0;
for i = 40: 10: 1000
% Set value for CO2
atmos.co2air = i / 0.7;
% Calculate photosynthesis
[flux] = LeafPhotosynthesis (physcon, atmos, leaf, flux);
% Save data for output
p = p + 1;
x1(p) = atmos.par;
x2(p) = flux.tleaf - physcon.tfrz;
x3(p) = atmos.co2air;
x4(p) = flux.ci / atmos.co2air;
x5(p) = flux.ac - flux.rd;
x6(p) = flux.aj - flux.rd;
x7(p) = flux.an;
end
% Plot data
plot(x3,x5,'b',x3,x6,'r',x3,x7,'g')
title('C_3 photosynthesis')
xlabel('c_i (\mumol mol^{-1})')
ylabel('Photosynthesis (\mumol CO_2 m^{-2} s^{-1})')
legend('A_c','A_j','A_n','Location','best')
% Output data and then clear variables from memory
A = [x1; x2; x3; x4; x5; x6; x7];
filename = 'co2_response.txt';
fileID = fopen(filename,'w');
fprintf(fileID,'%9.4f %9.4f %9.4f %9.4f %9.4f %9.4f %9.4f\n', A);
fclose(fileID);
clear x1 x2 x3 x4 x5 x6 x7 A;
% Reset to default value
atmos.co2air = co2conc;
end
% --- Temperature response at saturated light and standard CO2
if (plot_type == 3)
p = 0;
for i = 10: 1: 35
% Set value for leaf temperature
flux.tleaf = physcon.tfrz + i;
% Calculate photosynthesis
[flux] = LeafPhotosynthesis (physcon, atmos, leaf, flux);
% Save data for output
p = p + 1;
x1(p) = atmos.par;
x2(p) = flux.tleaf - physcon.tfrz;
x3(p) = atmos.co2air;
x4(p) = flux.ci / atmos.co2air;
x5(p) = flux.ac - flux.rd;
x6(p) = flux.aj - flux.rd;
x7(p) = flux.an;
end
% Plot data
plot(x2,x5,'b',x2,x6,'r',x2,x7,'g')
title('C_3 photosynthesis')
xlabel('Temperature (^{o}C)')
ylabel('Photosynthesis (\mumol CO_2 m^{-2} s^{-1})')
legend('A_c','A_j','A_n','Location','best')
% Output data and then clear variables from memory
A = [x1; x2; x3; x4; x5; x6; x7];
filename = 'temperature_response.txt';
fileID = fopen(filename,'w');
fprintf(fileID,'%9.4f %9.4f %9.4f %9.4f %9.4f %9.4f %9.4f\n', A);
fclose(fileID);
clear x1 x2 x3 x4 x5 x6 x7 A;
% Reset to default value
flux.tleaf = leaf_temp;
end
Aux. programs
LeafPhotosynthesis.m | View on GitHub
function [flux] = LeafPhotosynthesis (physcon, atmos, leaf, flux)
% Calculate leaf photosynthesis given a known Ci
% ------------------------------------------------------
% Input
% physcon.tfrz ! Freezing point of water (K)
% physcon.rgas ! Universal gas constant (J/K/mol)
% atmos.co2air ! Atmospheric CO2 (umol/mol)
% atmos.o2air ! Atmospheric O2 (mmol/mol)
% leaf.vcmax25 ! Maximum carboxylation rate at 25C (umol/m2/s)
% leaf.jmax25 ! Maximum electron transport rate at 25C (umol/m2/s)
% leaf.rd25 ! Leaf respiration rate at 25C (umol CO2/m2/s)
% leaf.kc25 ! Michaelis-Menten constant for CO2 at 25C (umol/mol)
% leaf.ko25 ! Michaelis-Menten constant for O2 at 25C (mmol/mol)
% leaf.cp25 ! CO2 compensation point at 25C (umol/mol)
% leaf.kcha ! Activation energy for Kc (J/mol)
% leaf.koha ! Activation energy for Ko (J/mol)
% leaf.cpha ! Activation energy for Cp (J/mol)
% leaf.vcmaxha ! Activation energy for Vcmax (J/mol)
% leaf.jmaxha ! Activation energy for Jmax (J/mol)
% leaf.rdha ! Activation energy for Rd (J/mol)
% leaf.vcmaxhd ! Deactivation energy for Vcmax (J/mol)
% leaf.jmaxhd ! Deactivation energy for Jmax (J/mol)
% leaf.rdhd ! Deactivation energy for Rd (J/mol)
% leaf.vcmaxse ! Entropy term for Vcmax (J/mol/K)
% leaf.jmaxse ! Entropy term for Jmax (J/mol/K)
% leaf.rdse ! Entropy term for Rd (J/mol/K)
% leaf.vcmaxc ! Vcmax scaling factor for high temperature inhibition (25 C = 1.0)
% leaf.jmaxc ! Jmax scaling factor for high temperature inhibition (25 C = 1.0)
% leaf.rdc ! Rd scaling factor for high temperature inhibition (25 C = 1.0)
% leaf.phi_psii ! Quantum yield of PS II
% leaf.theta_j ! Empirical curvature parameter for electron transport rate
% leaf.colim ! Photosynthesis co-limitation: 0 = no. 1 = yes
% leaf.colim_c3 ! Empirical curvature parameter for C3 co-limitation
% flux.apar ! Leaf absorbed PAR (umol photon/m2 leaf/s)
% flux.tleaf ! Leaf temperature (K)
%
% Output
% flux.vcmax ! Maximum carboxylation rate (umol/m2/s)
% flux.jmax ! Maximum electron transport rate (umol/m2/s)
% flux.cp ! CO2 compensation point (umol/mol)
% flux.kc ! Michaelis-Menten constant for CO2 (umol/mol)
% flux.ko ! Michaelis-Menten constant for O2 (mmol/mol)
% flux.je ! Electron transport rate (umol/m2/s)
% flux.ac ! Leaf Rubisco-limited gross photosynthesis (umol CO2/m2 leaf/s)
% flux.aj ! Leaf RuBP regeneration-limited gross photosynthesis (umol CO2/m2 leaf/s)
% flux.ag ! Leaf gross photosynthesis (umol CO2/m2 leaf/s)
% flux.an ! Leaf net photosynthesis (umol CO2/m2 leaf/s)
% flux.rd ! Leaf respiration rate (umol CO2/m2 leaf/s)
% flux.ci ! Leaf intercellular CO2 (umol/mol)
% ------------------------------------------------------
% --- Adjust photosynthetic parameters for temperature
ft = @(tl, ha) exp(ha/(physcon.rgas*(physcon.tfrz+25)) * (1-(physcon.tfrz+25)/tl));
fth = @(tl, hd, se, fc) fc / (1 + exp((-hd+se*tl)/(physcon.rgas*tl)));
flux.kc = leaf.kc25 * ft(flux.tleaf, leaf.kcha);
flux.ko = leaf.ko25 * ft(flux.tleaf, leaf.koha);
flux.cp = leaf.cp25 * ft(flux.tleaf, leaf.cpha);
t1 = ft(flux.tleaf, leaf.vcmaxha);
t2 = fth(flux.tleaf, leaf.vcmaxhd, leaf.vcmaxse, leaf.vcmaxc);
flux.vcmax = leaf.vcmax25 * t1 * t2;
t1 = ft(flux.tleaf, leaf.jmaxha);
t2 = fth(flux.tleaf, leaf.jmaxhd, leaf.jmaxse, leaf.jmaxc);
flux.jmax = leaf.jmax25 * t1 * t2;
t1 = ft(flux.tleaf, leaf.rdha);
t2 = fth(flux.tleaf, leaf.rdhd, leaf.rdse, leaf.rdc);
flux.rd = leaf.rd25 * t1 * t2;
% --- Electron transport rate for C3 plants
% Solve the polynomial: aquad*je^2 + bquad*je + cquad = 0
% for Je. Correct solution is the smallest of the two roots.
qabs = 0.5 * leaf.phi_psii * flux.apar;
aquad = leaf.theta_j;
bquad = -(qabs + flux.jmax);
cquad = qabs * flux.jmax;
pcoeff = [aquad bquad cquad];
proots = roots(pcoeff);
flux.je = min(proots(1), proots(2));
% --- Specify Ci
flux.ci = 0.7 * atmos.co2air;
% --- C3: Rubisco-limited photosynthesis
flux.ac = flux.vcmax * max(flux.ci - flux.cp, 0) / (flux.ci + flux.kc * (1 + atmos.o2air / flux.ko));
% --- C3: RuBP regeneration-limited photosynthesis
flux.aj = flux.je * max(flux.ci - flux.cp, 0) / (4 * flux.ci + 8 * flux.cp);
% --- Photosynthesis is the minimum or co-limited rate
if (leaf.colim == 1) % Use co-limitation
% Co-limit Ac and Aj. Ag is the co-limited photosynthesis rate found
% by solving the polynomial: aquad*Ag^2 + bquad*Ag + cquad = 0 for Ag.
% Correct solution is the smallest of the two roots.
aquad = leaf.colim_c3;
bquad = -(flux.ac + flux.aj);
cquad = flux.ac * flux.aj;
pcoeff = [aquad bquad cquad];
proots = roots(pcoeff);
flux.ag = min(proots(1), proots(2));
elseif (leaf.colim == 0) % No co-limitation
flux.ag = min(flux.ac, flux.aj);
end
% Net CO2 uptake
flux.an = flux.ag - flux.rd;
LeafPhysiologyParams.m | View on GitHub
function [leaf] = LeafPhysiologyParams (params, physcon, leaf)
% ------------------------------------------------------
% Input
% params.vis ! Waveband index for visible radiation
% physcon.tfrz ! Freezing point of water (K)
% physcon.rgas ! Universal gas constant (J/K/mol)
%
% Output
% leaf.vcmax25 ! Maximum carboxylation rate at 25C (umol/m2/s)
% leaf.jmax25 ! Maximum electron transport rate at 25C (umol/m2/s)
% leaf.rd25 ! Leaf respiration rate at 25C (umol CO2/m2/s)
% leaf.kc25 ! Michaelis-Menten constant for CO2 at 25C (umol/mol)
% leaf.ko25 ! Michaelis-Menten constant for O2 at 25C (mmol/mol)
% leaf.cp25 ! CO2 compensation point at 25C (umol/mol)
% leaf.kcha ! Activation energy for Kc (J/mol)
% leaf.koha ! Activation energy for Ko (J/mol)
% leaf.cpha ! Activation energy for Cp (J/mol)
% leaf.vcmaxha ! Activation energy for Vcmax (J/mol)
% leaf.jmaxha ! Activation energy for Jmax (J/mol)
% leaf.rdha ! Activation energy for Rd (J/mol)
% leaf.vcmaxhd ! Deactivation energy for Vcmax (J/mol)
% leaf.jmaxhd ! Deactivation energy for Jmax (J/mol)
% leaf.rdhd ! Deactivation energy for Rd (J/mol)
% leaf.vcmaxse ! Entropy term for Vcmax (J/mol/K)
% leaf.jmaxse ! Entropy term for Jmax (J/mol/K)
% leaf.rdse ! Entropy term for Rd (J/mol/K)
% leaf.vcmaxc ! Vcmax scaling factor for high temperature inhibition (25 C = 1.0)
% leaf.jmaxc ! Jmax scaling factor for high temperature inhibition (25 C = 1.0)
% leaf.rdc ! Rd scaling factor for high temperature inhibition (25 C = 1.0)
% leaf.phi_psii ! Quantum yield of PS II
% leaf.theta_j ! Empirical curvature parameter for electron transport rate
% leaf.colim_c3 ! Empirical curvature parameter for C3 co-limitation
% leaf.rho ! Leaf reflectance for visible (vis) and near-infrared (nir) wavebands
% leaf.tau ! Leaf transmittance for visible (vis) and near-infrared (nir) wavebands
% ------------------------------------------------------
% --- Vcmax and other parameters (at 25C)
leaf.vcmax25 = 60;
leaf.jmax25 = 1.67 * leaf.vcmax25;
leaf.rd25 = 0.015 * leaf.vcmax25;
% --- Kc, Ko, Cp at 25C
leaf.kc25 = 404.9;
leaf.ko25 = 278.4;
leaf.cp25 = 42.75;
% --- Activation energy
leaf.kcha = 79430;
leaf.koha = 36380;
leaf.cpha = 37830;
leaf.rdha = 46390;
leaf.vcmaxha = 65330;
leaf.jmaxha = 43540;
% --- High temperature deactivation
% Deactivation energy (J/mol)
leaf.rdhd = 150000;
leaf.vcmaxhd = 150000;
leaf.jmaxhd = 150000;
% Entropy term (J/mol/K)
leaf.rdse = 490;
leaf.vcmaxse = 490;
leaf.jmaxse = 490;
% Scaling factors for high temperature inhibition (25 C = 1.0).
% The factor "c" scales the deactivation to a value of 1.0 at 25C.
fth25 = @(hd, se) 1 + exp((-hd + se*(physcon.tfrz+25)) / (physcon.rgas*(physcon.tfrz+25)));
leaf.vcmaxc = fth25 (leaf.vcmaxhd, leaf.vcmaxse);
leaf.jmaxc = fth25 (leaf.jmaxhd, leaf.jmaxse);
leaf.rdc = fth25 (leaf.rdhd, leaf.rdse);
% --- C3 parameters
% Quantum yield of PS II
leaf.phi_psii = 0.85;
% Empirical curvature parameter for electron transport rate
leaf.theta_j = 0.90;
% Empirical curvature parameter for C3 co-limitation
leaf.colim_c3 = 0.98;
% --- Leaf reflectance and transmittance: visible and near-infrared wavebands
leaf.rho(params.vis) = 0.10;
leaf.tau(params.vis) = 0.10;
Output
Figures
Figure 1
Text
light_response.txt | View on GitHub | View raw