DAYCENT
Table of contents
Code
Main program
sp_18_01.m | View on GitHub
% Supplemental program 18.1
% This program simulates litter decomposition and soil organic matter dynamics using DAYCENT
% as described in Figure 18.8. The program follows carbon and nitrogen pools and fluxes
% associated with the addition of a single pulse of leaf litter on the surface or root litter
% belowground. This is the litter bag protocal in the simulations of Bonan et al.
% (2013) Global Change Biology 19:957-974. This is simplified code, to illustrate the basics
% of carbon and nitrogen dynamics. The full DAYCENT model is more complex and includes
% additional processes.
% --------------
% Initialization
% --------------
% --- number of carbon pools
npool = 12;
% pools:
% 1 = metabolic litter (SRFC)
% 2 = metabolic litter (SOIL)
% 3 = structural litter (SRFC)
% 4 = structural litter (SOIL)
% 5 = coarse woody debris: fine branch
% 6 = coarse woody debris: large wood
% 7 = coarse woody debris: coarse root
% 8 = active SOM1 (SRFC)
% 9 = active SOM1 (SOIL)
% 10 = slow SOM2 (SRFC)
% 11 = slow SOM2 (SOIL)
% 12 = passive SOM3
% --- number of litter fluxes
nlitflx = 5;
% 1 = leaf
% 2 = fine root
% 3 = coarse woody debris (fine branch)
% 4 = coarse woody debris (large wood)
% 5 = coarse woody debris (coarse root)
% --- specify biome type (for mixing of surface SOM2 to soil SOM2)
grass = 1;
forest = 2;
biome = grass;
% --- site conditions
sand = 50; % percent sand
clay = 20; % percent clay
cdi = 0.8; % soil temperature and moisture factor (0-1); used as a constant in this example
pH = 7.0; % pH
O2 = 1.0; % effect of soil anaerobic conditions on decomposition (0-1)
soilN = 3; % soil mineral nitrogen (gN/m2); determines C/N of SOM pools
% --- effect of cultivation on decomposition (1:SOM1, 2:SOM2, 3:SOM3, 4:structural)
% These are not used here. DAYCENT has factors specific to various cultivation types.
cultfac(1) = 1;
cultfac(2) = 1;
cultfac(3) = 1;
cultfac(4) = 1;
% --- coarse woody debris chemistry
cwdlig(1) = 0.25; % lignin fraction for fine branch
cwdlig(2) = 0.25; % lignin fraction for large wood
cwdlig(3) = 0.25; % lignin fraction for coarse root
% --- lignin
rsplig = 0.3; % fraction of carbon lost as respiration (lignin)
% --- leaf and fine root litter chemistry
% generic values (need to be specified for leaf and fine root)
leaf_flig = 0.159; % leaf litter lignin fraction
leaf_cn = 61.8; % leaf litter C:N (gC/gN)
froot_flig = 0.349; % fine root litter lignin fraction
froot_cn = 61.5; % fine root C:N (gC/gN)
% chose specific litter type for litterbag study
litter = 'TRAEf'; % leaf: Triticum aestivum
% litter = 'PIREf'; % leaf: Pinus resinosa
% litter = 'THPLf'; % leaf: Western red cedar
% litter = 'ACSAf'; % leaf: sugar maple
% litter = 'QUPRf'; % leaf: chestnut oak
% litter = 'DRGLf'; % leaf: tropical broadleaf
% litter = 'ANGEr'; % root: big bluestem grass
% litter = 'PIELr'; % root: slash pine
% litter = 'DRGLr'; % root: tropical broadleaf
% specific litter types
switch litter
case 'TRAEf' % leaf: Triticum aestivum
leaf_flig = 0.162; % leaf litter lignin fraction
leaf_cn = 133.3; % leaf litter C:N (gC/gN)
case 'PIREf' % leaf: Pinus resinosa
leaf_flig = 0.192; % leaf litter lignin fraction
leaf_cn = 92.7; % leaf litter C:N (gC/gN)
case 'THPLf' % leaf: Western red cedar
leaf_flig = 0.267; % leaf litter lignin fraction
leaf_cn = 83.1; % leaf litter C:N (gC/gN)
case 'ACSAf' % leaf: sugar maple
leaf_flig = 0.159; % leaf litter lignin fraction
leaf_cn = 61.8; % leaf litter C:N (gC/gN)
case 'QUPRf' % leaf: chestnut oak
leaf_flig = 0.235; % leaf litter lignin fraction
leaf_cn = 50.5; % leaf litter C:N (gC/gN)
case 'DRGLf' % leaf: tropical broadleaf
leaf_flig = 0.109; % leaf litter lignin fraction
leaf_cn = 24.2; % leaf litter C:N (gC/gN)
case 'ANGEr' % root: big bluestem grass
froot_flig = 0.105; % fine root litter lignin fraction
froot_cn = 59.4; % fine root C:N (gC/gN)
case 'PIELr' % root: slash pine
froot_flig = 0.349; % fine root litter lignin fraction
froot_cn = 61.5; % fine root C:N (gC/gN)
case 'DRGLr' % root: tropical broadleaf
froot_flig = 0.161; % fine root litter lignin fraction
froot_cn = 64.6; % fine root C:N (gC/gN)
end
% --- litter flux - U: 1=leaf, 2=fine root, 3=cwd (fine branch), 4=cwd (large wood), 5=cwd (coarse root)
% DAYCENT: fraction of large wood CWD flux that is fine branch
fbranch = 0.10;
% fluxes: gC/m2/year -> gC/m2/sec
U(1,1) = 0 / (365 * 86400);
U(2,1) = 0 / (365 * 86400);
U(3,1) = 0 / (365 * 86400) * fbranch;
U(4,1) = 0 / (365 * 86400) * (1 - fbranch);
U(5,1) = 0 / (365 * 86400);
% C/N ratios of litter fluxes
U_cn(1) = leaf_cn;
U_cn(2) = froot_cn;
U_cn(3) = 500;
U_cn(4) = 500;
U_cn(5) = 500;
% --- initial pool size
% C(i,1) = carbon for pool i (g C/m2)
% N(i,1) = nitrogen for pool i (g N/m2)
C = zeros(npool,1);
N = zeros(npool,1);
% ------------------
% Time stepping loop
% ------------------
% --- length of time step (seconds)
dt = 1800; % 30-minutes
% --- number of timsteps per day
ntim = 86400 / dt;
% --- number of years to simulate
nyears = 11;
% --- days per month
ndays = [31, 28, 31, 30, 31, 30, 31, 31, 30, 31, 30, 31];
% --- initialize step counter
nstep = 0;
% --- counter for daily output
ncount = 0;
% --- advance time
for year = 1:nyears
for month = 1:12
for day = 1:ndays(month)
for itim = 1:ntim
nstep = nstep + 1; % step counter
cumday = nstep / ntim; % cumulative day
cumyear = nstep / (ntim * 365); % cumulative year
if (month == 1 && day == 1 && itim == 1)
fprintf('year = %8.1f\n',cumyear)
end
if (itim == 1 && day == 1)
fprintf('day = %6.0f\n',cumday)
end
% =============================
% add litter on first time step
% =============================
if (litter == 'TRAEf' | litter == 'PIREf' | litter == 'THPLf' | ...
litter == 'ACSAf' | litter == 'QUPRf' | litter == 'DRGLf')
add_leaf = 100;
add_root = 0;
end
if (litter == 'ANGEr' | litter == 'PIELr' | litter == 'DRGLr')
add_leaf = 0;
add_root = 100;
end
if (nstep == 1)
U(1,1) = add_leaf / dt;
U(2,1) = add_root / dt;
U(3,1) = 0;
U(4,1) = 0;
U(5,1) = 0;
litterC = add_leaf + add_root; % save litter C input
litterN = add_leaf / leaf_cn + add_root / froot_cn; % save litter N input
else
U(1,1) = 0;
U(2,1) = 0;
U(3,1) = 0;
U(4,1) = 0;
U(5,1) = 0;
end
% ===============================================
% calculate carbon fluxes and update carbon pools
% ===============================================
% matrix B to partition litter fluxes to each carbon pool
% and also lignin fraction of structural litter strlig
[strlig, B] = litter_partition_matrix (leaf_flig, leaf_cn, froot_flig, froot_cn);
% base decomposition rate K and mixing Kmix
[K, Kmix] = decomp_rate_base (biome, grass, forest);
% environmental scalar xi adjusts base decomposition rate
[K_s21, xi] = decomp_rate_scalar (cdi, pH, O2, sand, strlig, cwdlig, cultfac, K, Kmix);
% carbon transfer matrix A
% pathf = fractional carbon flow from pool j to pool i
% respf = fractional respiration loss for carbon flow from pool j to pool i
[A, pathf, respf] = carbon_transfer_matrix (npool, sand, clay, O2, strlig, cwdlig, rsplig, Kmix, K_s21);
% calculate pool increment dC for each pool i - this
% is the generalized calculation
for i = 1:npool
dC(i,1) = 0;
% litter flux input
for j = 1:nlitflx
dC(i,1) = dC(i,1) + B(i,j) * U(j,1);
end
% carbon transfer from pool j to pool i
for j = 1:npool
if (j ~= i)
dC(i,1) = dC(i,1) + (1 - respf(i,j)) * pathf(i,j) * xi(j,j) * K(j,j) * C(j,1);
end
end
% carbon loss from pool i
dC(i,1) = dC(i,1) - xi(i,i) * K(i,i) * C(i,1);
end
% ... or use matrix algebra: dC = B * U + A * xi * K * C
% dC = B * U + A * xi * K * C;
% heterotrophic respiration
RH = 0;
for i = 1:npool
for j = 1:npool
if (j ~= i)
RH = RH + respf(i,j) * pathf(i,j) * xi(j,j) * K(j,j) * C(j,1);
end
end
end
% =====================================
% calculate N fluxes and update N pools
% =====================================
% specify C/N ratio of SOM pools based on soil mineral N
cnrat = @(x,cnmin,cnmax,ncrit) max(min(((1-x/ncrit)*(cnmax-cnmin)+cnmin),cnmax),cnmin);
for i = 1:7
som_cn(i) = 0;
end
som_cn( 8) = cnrat(soilN, 10, 20, 1);
som_cn( 9) = cnrat(soilN, 8, 18, 2);
som_cn(10) = cnrat(soilN, 12, 15, 2);
som_cn(11) = cnrat(soilN, 12, 40, 2);
som_cn(12) = cnrat(soilN, 6, 20, 2);
% N input from litter flux j to pool i. note the
% special case for leaf and fine root litter
% to the structural pool (which has C/N = 200).
N_in_total = U(1,1) / U_cn(1);
N_to_struc = B(3,1) * U(1,1) / 200;
N_to_metab = N_in_total - N_to_struc;
N_litter_flux(1) = N_to_metab;
N_litter_flux(3) = N_to_struc;
N_in_total = U(2,1) / U_cn(2);
N_to_struc = B(4,2) * U(2,1) / 200;
N_to_metab = N_in_total - N_to_struc;
N_litter_flux(2) = N_to_metab;
N_litter_flux(4) = N_to_struc;
N_litter_flux(5) = B(5,3) * U(3,1) / U_cn(3);
N_litter_flux(6) = B(6,4) * U(4,1) / U_cn(4);
N_litter_flux(7) = B(7,5) * U(5,1) / U_cn(5);
N_litter_flux(8) = 0;
N_litter_flux(9) = 0;
N_litter_flux(10) = 0;
N_litter_flux(11) = 0;
N_litter_flux(12) = 0;
% N flow in carbon transfer among pools
for i = 1:npool
N_transfer(i) = 0;
% N transfer from donor pool j to receiver pool i
for j = 1:npool
if (j ~= i)
N_transfer(i) = N_transfer(i) + pathf(i,j) * xi(j,j) * K(j,j) * N(j,1);
end
end
% N loss from pool i
N_transfer(i) = N_transfer(i) - xi(i,i) * K(i,i) * N(i,1);
end
% mineral N flux in carbon transfer from donor pool j to SOM receiver pool i
for i = 1:12
N_mineral_flux(i) = 0;
end
for i = 8:12
for j = 1:npool
% carbon flow from pool j to pool i
tcflow = pathf(i,j) * xi(j,j) * K(j,j) * C(j,1);
if (tcflow > 0)
% C/N ratio of donor pool
cn_donor = C(j,1) / N(j,1);
% C/N ratio of receiver SOM pool
cn_receiver = som_cn(i);
% special case for SOM2(SRFC) -> SOM2(SOIL); there is no immobilization
% or mineralization because this is a mixing flux
if (j == 10 & i == 11)
cn_receiver = cn_donor;
end
% Note the special case when lignin decomposes to SOM1 and SOM2. In the
% DAYCENT code used by Bonan et al. (2013), the receiver C/N ratios are:
% SOM1: cn_receiver = cnrat(soilN, 10, 20, 1)
% SOM2: cn_receiver = cnrat(soilN, 12, 15, 2)
% and do not vary between SRFC and SOIL pools. This means that the target
% C/N for SOIL pools is the same as for SRFC pools.
% special case when lignin decomposes to SOM1(SOIL): use values for SOM1(SRFC)
if ((j == 4 & i == 9) | (j == 7 & i == 9))
cn_receiver = cnrat(soilN, 10, 20, 1);
end
% special case when lignin decomposes to SOM2(SOIL): use values for SOM2(SRFC)
if ((j == 4 & i == 11) | (j == 7 & i == 11))
cn_receiver = cnrat(soilN, 12, 15, 2);
end
% mineral N flux for flow from donor to receiver (+ = immobilization)
if (j ~= i)
N_flux = tcflow * ((1 - respf(i,j)) / cn_receiver - 1/cn_donor);
end
% sum N flux to pool i
N_mineral_flux(i) = N_mineral_flux(i) + N_flux;
end
end
end
% sum N fluxes for each pool i
for i = 1:npool
dN(i,1) = N_litter_flux(i) + N_transfer(i) + N_mineral_flux(i);
end
% ============
% update pools
% ============
% carbon pools
for i = 1:npool
C(i,1) = C(i,1) + dC(i,1) * dt;
end
litC = C(1,1) + C(2,1) + C(3,1) + C(4,1); % litter: metabolic + structural
cwdC = C(5,1) + C(6,1) + C(7,1); % coarse woody debris
somC = C(8,1) + C(9,1) + C(10,1) + C(11,1) + C(12,1); % soil organic matter
totC = litC + cwdC + somC; % total carbon
% balance check
dCtot = 0;
for i = 1:npool
dCtot = dCtot + dC(i,1);
end
Utot = 0;
for i = 1:nlitflx
Utot = Utot + U(i,1);
end
err = (dCtot - (Utot - RH)) * dt;
if (abs(err) > 1e-12)
fprintf('err = %15.5f\n',err)
error ('CARBON BALANCE ERROR')
end
% nitrogen pools
totN_old = 0;
for i = 1:npool
totN_old = totN_old + N(i,1);
end
for i = 1:npool
N(i,1) = N(i,1) + dN(i,1) * dt;
end
litN = N(1,1) + N(2,1) + N(3,1) + N(4,1); % litter: metabolic + structural
cwdN = N(5,1) + N(6,1) + N(7,1); % coarse woody debris
somN = N(8,1) + N(9,1) + N(10,1) + N(11,1) + N(12,1); % soil organic matter
totN = litN + cwdN + somN; % total nitrogen
% balance check
Utot = 0;
Nmin_tot = 0;
for i = 1:npool
Utot = Utot + N_litter_flux(i);
Nmin_tot = Nmin_tot + N_mineral_flux(i);
end
dNtot = totN - totN_old;
err = dNtot - (Utot + Nmin_tot) * dt;
if (abs(err) > 1e-12)
fprintf('err = %15.5f\n',err)
error ('NITROGEN BALANCE ERROR')
end
% save daily output for graphing
if (itim == 1)
ncount = ncount + 1;
x1(ncount) = cumyear;
y1(ncount) = totC;
y2(ncount) = totN;
end
end
end
end
end
% ----------------------
% graph data
% ----------------------
% plot C and N vs time
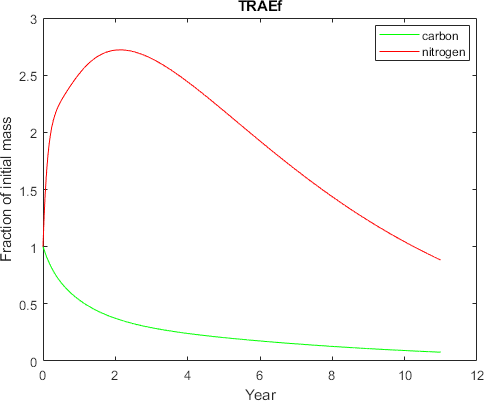
plot(x1,y1/litterC,'g-',x1,y2/litterN,'r-')
xlabel('Year')
ylabel('Fraction of initial mass')
legend('carbon','nitrogen','Location','best')
title(litter)
% plot N vs C
% plot(y1/litterC,y2/litterN)
% set(gca, 'XDir','reverse')
% xlabel('Mass remaining (fraction)')
% ylabel('Fraction of initial nitrogen')
% title(litter)
% ----------------------------------
% Write formated output to text file
% ----------------------------------
data = [x1; y1; y2];
fileID = fopen('data.txt','w');
fprintf(fileID,'%15s %15s %15s\n','year','carbon','nitrogen');
fprintf(fileID,'%15.5f %15.5f %15.5f\n',data);
Aux. programs
carbon_transfer_matrix.m | View on GitHub
function [A, pathf, respf] = carbon_transfer_matrix (npool, sand, clay, O2, strlig, cwdlig, rsplig, Kmix, K_s21)
% Carbon transfer matrix
% ---------------------------------------------------------------------------------------------
% Input
% npool ! number of carbon pools
% sand ! percent sand
% clay ! percent clay
% O2 ! effect of soil anaerobic conditions on decomposition (0-1)
% strlig ! lignin fraction: (1) SRFC and (2) SOIL structural litter (g lignin/g biomass)
% cwdlig ! lignin fraction: (1) fine branch; (2) large wood; (3) coarse root
% rsplig ! fraction of carbon lost as respiration (lignin)
% Kmix ! base mixing rate: SOM2(SRFC) -> SOM2(SOIL), 1/sec
% K_s21 ! rate constant: total loss from SOM2(SRFC), 1/sec
%
% Output
% A ! carbon transfer matrix
% pathf ! fractional carbon flow from pool j to pool i
% respf ! fractional respiration loss for carbon flow from pool j to pool i
% ---------------------------------------------------------------------------------------------
% --- zero out arrays
for i = 1:npool
for j = 1:npool
pathf(i,j) = 0;
respf(i,j) = 0;
A(i,j) = 0;
end
end
% --- anaerobic factor
fanerb = 1 + 5 * (1 - O2);
% --- pathf(i,j) = fractional carbon flow from pool j to pool i
pathf(8,1) = 1;
pathf(9,2) = 1;
pathf(8,3) = 1 - strlig(1);
pathf(10,3) = strlig(1);
pathf(9,4) = 1 - strlig(2);
pathf(11,4) = strlig(2);
pathf(8,5) = 1 - cwdlig(1);
pathf(10,5) = cwdlig(1);
pathf(8,6) = 1 - cwdlig(2);
pathf(10,6) = cwdlig(2);
pathf(9,7) = 1 - cwdlig(3);
pathf(11,7) = cwdlig(3);
pathf(10,8) = 1;
pathf(12,9) = (0.003 + 0.032 * clay/100) * fanerb;
pathf(11,9) = 1 - pathf(12,9);
pathf(11,10) = Kmix / K_s21;
pathf(8,10) = 1 - pathf(11,10);
pathf(12,11) = (0.003 + 0.009 * clay/100) * fanerb;
pathf(9,11) = 1 - pathf(12,11);
pathf(9,12) = 1;
% --- respf(i,j) = fractional respiration loss for carbon flow from pool j to pool i
respf(8,1) = 0.55;
respf(9,2) = 0.55;
respf(8,3) = 0.45;
respf(10,3) = rsplig;
respf(9,4) = 0.55;
respf(11,4) = rsplig;
respf(8,5) = 0.45;
respf(10,5) = rsplig;
respf(8,6) = 0.45;
respf(10,6) = rsplig;
respf(9,7) = 0.55;
respf(11,7) = rsplig;
respf(10,8) = 0.60;
respf(12,9) = 0;
respf(11,9) = (0.17 + 0.68 * sand/100) / pathf(11,9);
respf(11,10) = 0;
respf(8,10) = 0.55;
respf(12,11) = 0;
respf(9,11) = 0.55 / pathf(9,11);
respf(9,12) = 0.55 * O2;
% --- carbon transfer matrix: A(i,j) = fractional carbon flow from pool j that enters pool i
for i = 1:npool
A(i,i) = -1;
end
for i = 1:npool
for j = 1:npool
if (j ~= i)
A(i,j) = pathf(i,j) * (1 - respf(i,j));
end
end
end
decomp_rate_base.m | View on GitHub
function [K, Kmix] = decomp_rate_base (biome, grass, forest)
% -----------------------------------------------------
% Base decomposition rate for each carbon pool (per sec)
% -----------------------------------------------------
% convert rate from per year to per second
K(1,1) = 8.0 / (365 * 86400);
K(2,2) = 18.5 / (365 * 86400);
K(3,3) = 2.0 / (365 * 86400);
K(4,4) = 4.9 / (365 * 86400);
K(5,5) = 1.5 / (365 * 86400);
K(6,6) = 0.02 / (365 * 86400);
K(7,7) = 0.1 / (365 * 86400);
K(8,8) = 6.0 / (365 * 86400);
K(9,9) = 11.0 / (365 * 86400);
K(10,10) = 0.08 / (365 * 86400);
K(11,11) = 0.4 / (365 * 86400);
K(12,12) = 0.0033 / (365 * 86400);
% mixing of som2c(SRFC) to som2c(SOIL)
% annual turnover time for grass/crop: 4 years
% annual turnover time for forest: 10 years
if (biome == grass)
Kmix = 0.25 / (365 * 86400);
elseif (biome == forest)
Kmix = 0.10 / (365 * 86400);
else
Kmix = 0;
end
decomp_rate_scalar.m | View on GitHub
function [K_s21, xi] = decomp_rate_scalar (cdi, pH, O2, sand, strlig, cwdlig, cultfac, K, Kmix)
% Environmental scalar for each carbon pool adjusts base
% rate for soil temperature and soil moisture scalars
% (cdi) and additionally pH, lignin, texture, anaerobic,
% and cultivation
% ----------------------------------------------------------------
% Input
% cdi ! soil temperature and moisture scalar (0-1)
% pH ! pH
% O2 ! effect of soil anaerobic conditions on decomposition (0-1)
% sand ! percent sand
% strlig ! lignin fraction: (1) SRFC and (2) SOIL structural litter (g lignin/g biomass)
% cwdlig ! lignin fraction: (1) fine branch; (2) large wood; (3) coarse root
% cultfac ! effect of cultivation on decomposition (1:SOM1, 2:SOM2, 3:SOM3, 4:structural)
% K ! base decomposition rate (1/sec)
% Kmix ! base mixing rate: SOM2(SRFC) -> SOM2(SOIL), 1/sec
%
% Output
% K_s21 ! rate constant: total loss from SOM2(SRFC), 1/sec
% xi ! environmental scalar
% ----------------------------------------------------------------
% function for pH effect
pH_factor = @(x,a,b,c,d) b + (c / pi) * atan(d * (x - a) * pi);
% ==========================================
% metabolic litter (SRFC)
% ==========================================
pHeff = pH_factor(pH, 4.8, 0.5, 1.14, 0.7);
pHeff = max(min(pHeff, 1), 0);
xi(1,1) = cdi * pHeff;
% ==========================================
% metabolic litter (SOIL)
% ==========================================
pHeff = pH_factor(pH, 4.8, 0.5, 1.14, 0.7);
pHeff = max(min(pHeff, 1), 0);
xi(2,2) = cdi * pHeff * O2;
% ==========================================
% structural litter (SRFC)
% ==========================================
pHeff = pH_factor(pH, 4.0, 0.5, 1.1, 0.7);
pHeff = max(min(pHeff, 1), 0);
xi(3,3) = cdi * pHeff * exp(-3*strlig(1));
% ==========================================
% structural litter (SOIL)
% ==========================================
pHeff = pH_factor(pH, 4.0, 0.5, 1.1, 0.7);
pHeff = max(min(pHeff, 1), 0);
xi(4,4) = cdi * pHeff * exp(-3*strlig(2)) * O2 * cultfac(4);
% ==========================================
% coarse woody debris: fine branch
% ==========================================
pHeff = pH_factor(pH, 4.0, 0.5, 1.1, 0.7);
pHeff = max(min(pHeff, 1), 0);
xi(5,5) = cdi * pHeff * exp(-3*cwdlig(1));
% ==========================================
% coarse woody debris: large wood
% ==========================================
pHeff = pH_factor(pH, 4.0, 0.5, 1.1, 0.7);
pHeff = max(min(pHeff, 1), 0);
xi(6,6) = cdi * pHeff * exp(-3*cwdlig(2));
% ==========================================
% coarse woody debris: coarse root
% ==========================================
pHeff = pH_factor(pH, 4.0, 0.5, 1.1, 0.7);
pHeff = max(min(pHeff, 1), 0);
xi(7,7) = cdi * pHeff * exp(-3*cwdlig(3)) * O2;
% ==========================================
% active soil organic matter: SOM1 (SRFC)
% ==========================================
pHeff = pH_factor(pH, 4.0, 0.5, 1.1, 0.7);
pHeff = max(min(pHeff, 1), 0);
xi(8,8) = cdi * pHeff;
% ==========================================
% active soil organic matter: SOM1 (SOIL)
% ==========================================
pHeff = pH_factor(pH, 4.8, 0.5, 1.14, 0.7);
pHeff = max(min(pHeff, 1), 0);
texture = 0.25 + 0.75 * (sand/100);
xi(9,9) = cdi * pHeff * O2 * texture * cultfac(1);
% ==========================================
% slow soil organic matter: SOM2 (SRFC)
% ==========================================
% som2(SRFC) -> som1(SRFC)
pHeff = pH_factor(pH, 4.0, 0.5, 1.1, 0.7);
pHeff = max(min(pHeff, 1), 0);
K_s21_to_s11 = K(10,10) * pHeff;
% som2(SRFC) -> som2(SOIL): mixing
K_s21_to_s22 = Kmix;
% total loss from som2(SRFC)
K_s21 = K_s21_to_s11 + K_s21_to_s22;
% effective environmental scalar
xi(10,10) = cdi * (K_s21 / K(10,10));
% ==========================================
% slow soil organic matter: SOM2 (SOIL)
% ==========================================
pHeff = pH_factor(pH, 4.0, 0.5, 1.1, 0.7);
pHeff = max(min(pHeff, 1), 0);
xi(11,11) = cdi * pHeff * O2 * cultfac(2);
% ==========================================
% passive soil organic matter: SOM3
% ==========================================
pHeff = pH_factor(pH, 3.0, 0.5, 1.1, 0.7);
pHeff = max(min(pHeff, 1), 0);
xi(12,12) = cdi * pHeff * O2 * cultfac(3);
litter_partition_matrix.m | View on GitHub
function [strlig, B] = litter_partition_matrix (leaf_flig, leaf_cn, froot_flig, froot_cn)
% Matrix to partition litter fluxes to each carbon pool
% ---------------------------------------------------------------------------------------------
% Input
% leaf_flig ! leaf litter lignin fraction
% leaf_cn ! leaf litter C:N (gC/gN)
% froot_flig ! fine root litter lignin fraction
% froot_cn ! fine root C:N (gC/gN)
%
% Output
% strlig ! lignin fraction: (1) SRFC and (2) SOIL structural litter (g lignin/g biomass)
% B ! litter flux partitioning matrix
% ---------------------------------------------------------------------------------------------
% --- leaf
% fraction of plant residue that is nitrogen (g N / g biomass)
fnit = 1 / (leaf_cn * 2.5);
% lignin fraction of plant residue (g lignin / g biomass)
flig = leaf_flig;
% lignin/nitrogen ratio of plant residue
rlig2n = flig / fnit;
% fraction of plant residue that goes to metabolic litter pool
fmet = 0.85 - 0.013 * rlig2n;
% make sure the fraction of residue which is lignin is not greater than the
% fraction which goes to structural
if (flig > (1 - fmet))
fmet = (1 - flig);
end
% minimum metabolic fraction
if (fmet < 0.20)
fmet = 0.20;
end
% adjust lignin content of structural litter pool. fligst is the fraction of
% incoming structural residue that is lignin
fligst = min(flig/(1 - fmet), 1);
% DAYCENT adjusts lignin content of structural litter pool for new material
% that is added. That is not needed in this program because only one type
% of leaf litter is added. If different types of leaf litter are added,
% need to adjust structural litter lignin
strlig(1) = fligst;
% B(i,j) = litter flux partitioning matrix (litter flux j -> pool i)
B( 1,1) = fmet;
B( 2,1) = 0;
B( 3,1) = 1 - fmet;
B( 4,1) = 0;
B( 5,1) = 0;
B( 6,1) = 0;
B( 7,1) = 0;
B( 8,1) = 0;
B( 9,1) = 0;
B(10,1) = 0;
B(11,1) = 0;
B(12,1) = 0;
% --- fine root
% fraction of plant residue that is nitrogen (g N / g biomass)
fnit = 1 / (froot_cn * 2.5);
% lignin fraction of plant residue (g lignin / g biomass)
flig = froot_flig;
% lignin/nitrogen ratio of plant residue
rlig2n = flig / fnit;
% fraction of plant residue that goes to metabolic litter pool
fmet = 0.85 - 0.013 * rlig2n;
% make sure the fraction of residue which is lignin is not greater than the
% fraction which goes to structural
if (flig > (1 - fmet))
fmet = (1 - flig);
end
% minimum metabolic fraction
if (fmet < 0.20)
fmet = 0.20;
end
% adjust lignin content of structural litter pool. fligst is the fraction of
% incoming structural residue that is lignin
fligst = min(flig/(1 - fmet), 1);
% DAYCENT adjusts lignin content of structural litter pool for new material
% that is added. That is not needed in this program because only one type
% of fine root litter is added. If different types of fine root litter are
% added, need to adjust structural litter lignin
strlig(2) = fligst;
% B(i,j) = litter flux partitioning matrix (litter flux j -> pool i)
B( 1,2) = 0;
B( 2,2) = fmet;
B( 3,2) = 0;
B( 4,2) = 1 - fmet;
B( 5,2) = 0;
B( 6,2) = 0;
B( 7,2) = 0;
B( 8,2) = 0;
B( 9,2) = 0;
B(10,2) = 0;
B(11,2) = 0;
B(12,2) = 0;
% --- cwd (fine branch)
B( 1,3) = 0;
B( 2,3) = 0;
B( 3,3) = 0;
B( 4,3) = 0;
B( 5,3) = 1;
B( 6,3) = 0;
B( 7,3) = 0;
B( 8,3) = 0;
B( 9,3) = 0;
B(10,3) = 0;
B(11,3) = 0;
B(12,3) = 0;
% cwd (large wood)
B( 1,4) = 0;
B( 2,4) = 0;
B( 3,4) = 0;
B( 4,4) = 0;
B( 5,4) = 0;
B( 6,4) = 1;
B( 7,4) = 0;
B( 8,4) = 0;
B( 9,4) = 0;
B(10,4) = 0;
B(11,4) = 0;
B(12,4) = 0;
% cwd (coarse root)
B( 1,5) = 0;
B( 2,5) = 0;
B( 3,5) = 0;
B( 4,5) = 0;
B( 5,5) = 0;
B( 6,5) = 0;
B( 7,5) = 1;
B( 8,5) = 0;
B( 9,5) = 0;
B(10,5) = 0;
B(11,5) = 0;
B(12,5) = 0;
Output
Figures
Figure 1
Text
data.txt | View on GitHub | View raw
sp_18_01_out.txt (standard output) | View on GitHub | View raw
year = 0.0
day = 0
day = 31
day = 59
day = 90
day = 120
day = 151
day = 181
day = 212
day = 243
day = 273
day = 304
day = 334
year = 1.0
day = 365
day = 396
day = 424
day = 455
day = 485
day = 516
day = 546
day = 577
day = 608
day = 638
day = 669
day = 699
year = 2.0
day = 730
day = 761
day = 789
day = 820
day = 850
day = 881
day = 911
day = 942
day = 973
day = 1003
day = 1034
day = 1064
year = 3.0
day = 1095
day = 1126
day = 1154
day = 1185
day = 1215
day = 1246
day = 1276
day = 1307
day = 1338
day = 1368
day = 1399
day = 1429
year = 4.0
day = 1460
day = 1491
day = 1519
day = 1550
day = 1580
day = 1611
day = 1641
day = 1672
day = 1703
day = 1733
day = 1764
day = 1794
year = 5.0
day = 1825
day = 1856
day = 1884
day = 1915
day = 1945
day = 1976
day = 2006
day = 2037
day = 2068
day = 2098
day = 2129
day = 2159
year = 6.0
day = 2190
day = 2221
day = 2249
day = 2280
day = 2310
day = 2341
day = 2371
day = 2402
day = 2433
day = 2463
day = 2494
day = 2524
year = 7.0
day = 2555
day = 2586
day = 2614
day = 2645
day = 2675
day = 2706
day = 2736
day = 2767
day = 2798
day = 2828
day = 2859
day = 2889
year = 8.0
day = 2920
day = 2951
day = 2979
day = 3010
day = 3040
day = 3071
day = 3101
day = 3132
day = 3163
day = 3193
day = 3224
day = 3254
year = 9.0
day = 3285
day = 3316
day = 3344
day = 3375
day = 3405
day = 3436
day = 3466
day = 3497
day = 3528
day = 3558
day = 3589
day = 3619
year = 10.0
day = 3650
day = 3681
day = 3709
day = 3740
day = 3770
day = 3801
day = 3831
day = 3862
day = 3893
day = 3923
day = 3954
day = 3984